Table Of Contents:
Scaling computational methodology for BeginNGS research
The vital role of federated queries in rare disease treatment
How TileDB is scaling federated queries across the BeginNGS consortium
Scaling computational methodology for BeginNGS research
Rare genetic conditions affect an estimated 300 million people worldwide and are the leading cause of child mortality and disability in high-income countries. This makes it crucial to test newborns for genetic diseases hours after birth to ensure timely diagnosis and treatment. However, the complexity of accurately performing this genome-based newborn screening (gNBS) at scale has been immense—slowing the diagnosis of severe, childhood-onset genetic diseases (SCGD) in the vital early days of life.
The BeginNGS platform was created to address this challenge. Using a combination of human expertise and artificial intelligence tools, BeginNGS is developing a list of actionable genetic disorders, associated interventions, target genes, and variants of interest, and a blocklist to minimize false positives in a newborn screening context. . The rapid whole genome sequencing (rWGS) approach being piloted by BeginNGS is meant to complement existing biochemical screens that have been employed for decades. . Testing of over 3,000 children with suspected genetic diseases revealed that 1 in 14 would have benefited from BeginNGS and would have received diagnoses and treatment 121 days earlier than testing after symptoms appeared.
The vital role of federated queries in rare disease treatment
One example of how this works was BeginNGS using genomic data from UK Biobank provided by Alexion to query alleles for rare diseases in a federated query. This enabled the BeginNGS team to make an efficient list of variants in target genes associated with actionable conditions, diseases for which an intervention exists, to scale up newborn screening worldwide, the addition of variants found in healthy adult populations to a blocklist achieved a 97 percent reduction in false positives. If the world’s newborn screening projects were to use federated queries to share genomic data across the planet, it would have a huge positive impact on the health outcomes for infants in NICUs everywhere.
How TileDB is scaling federated queries across the BeginNGS consortium
TileDB is proud to be a database technology partner for BeginNGS, lead by Rady Children's Institute for Genomic Medicine (RCIGM) at the newly formed Rady Children’s Health supporting their work in the treatment of rare diseases in newborns. We served as the variant warehouse and trusted research environment for the BeginNGS organization, who regularly ingest Variant Call Format (VCF) samples to their existing populations to identify 388 additional genetic diseases. The Rady team chose TileDB to handle their VCF samples in a 3-dimensional array on Amazon S3 using the TileDB-VCF open library, making this data analysis-ready on cloud storage.
Here’s an overview of how TileDB enables federated queries for BeginNGS by protecting sensitive genomic data with a limited user-defined function (UDF).
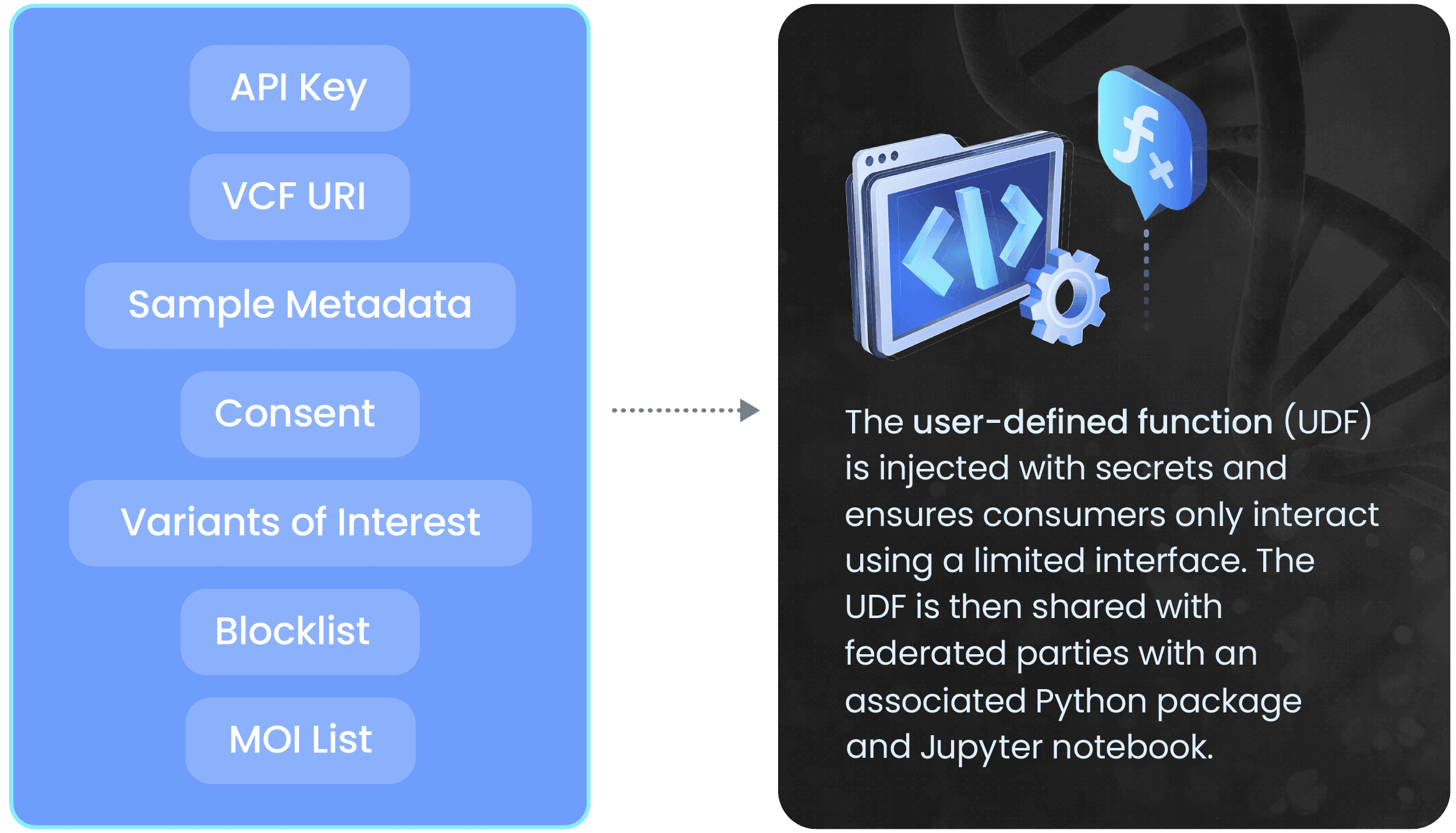
(currently BeginNGS v2, 53,855 P and LP variants that map to 342 genes, 412 SCGD and 1,603 SCGD therapeutic interventions) are normally encapsulated as a fixed resource in the UDF, but can be implemented as a parameter. This is pre-annotated with consequence and population frequency information, but only chr-pos-ref-alt is used for the query itself.
- these are entries BeginNGS classifies as NSDCC (non-severe disease causing in childhood).
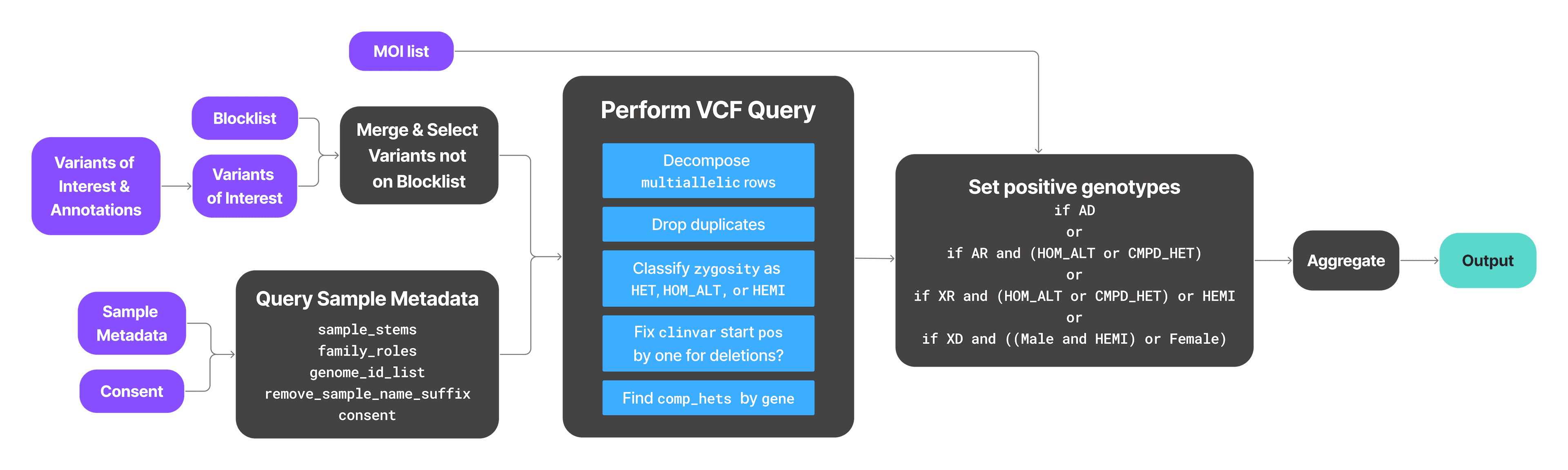
While the TileDB platform greatly simplifies the federated query process, here are the high-level steps if a non-TileDB user were to implement federated queries:
- 1
Apply blocklist to variants of interest (if using the recommended blocklist to screen out NSDCC entries).
- 2
Obtain VCF genotypes for variants of interest, then merge on chr,pos,ref,alt.
- 3
Classify compound hets in order to look for co-occurring hets in sample/gene groups.
- 4
Merge sample metadata to get sex, restricting use to only consented subjects.
- 5
Compute positive_genotypes using MOI rules.
- 6
For each gene/subject grouping, compose a concatenated string of observed hit variants (diplotypes).
Today, TileDB helps harmonize federated queries across a wide variety of newborn and healthy adult genomic datasets. This enables data owners across BeginNGS to write and distribute complex queries to consumers in other private namespaces that return aggregate results across all samples. Through TileDB’s expansion of the BeginNGS consortium’s federated query capabilities, we are enabling faster and more comprehensive analysis of variant datasets without compromising patient privacy. This results in quicker and more reliable answers to urgent genetic questions in the critical early days of life.
To learn more about how TileDB is scaling federated queries for more effective rare disease treatment, read the full case study on Rady Children’s Hospital.
About the author

Devika Garg
Head of Marketing
Devika Garg leads product marketing for life sciences at TileDB. Prior to TileDB, she ran marketing engines at Pure Storage, Proteus Digital Health and Applied Materials. A scientist and a science journalist in her past life, she loves to geek out on the latest discoveries and inventions. She earned her PhD at the National University of Singapore, her MS in Science Communication at University of California Santa Cruz, and her B Tech at the Indian Institute of Technology Kanpur.
Meet the authors

