A modern data platform for the next generation of single-cell biology
Scale your work on TileDB Cloud and maintain compatibility with existing tools.

TileDB's performant, scalable and cloud-native solution is ideal for our efforts at CZI to assemble and distribute one of the largest repositories of single-cell biological data in the world
Supercharge your multi-omics analysis

Unify single-cell data
Store data once as TileDB arrays, and enjoy interoperability with popular tools. We partnered with the Chan Zuckerberg Initiative to develop a data model that brings together the single-cell analysis toolchain.

Work at atlas scale
Parallelize complex workloads and create distributed algorithms using TileDB Cloud. Access policies allow your work to stay within your organization, or securely share it with external collaborators to unlock atlas-scale analysis.
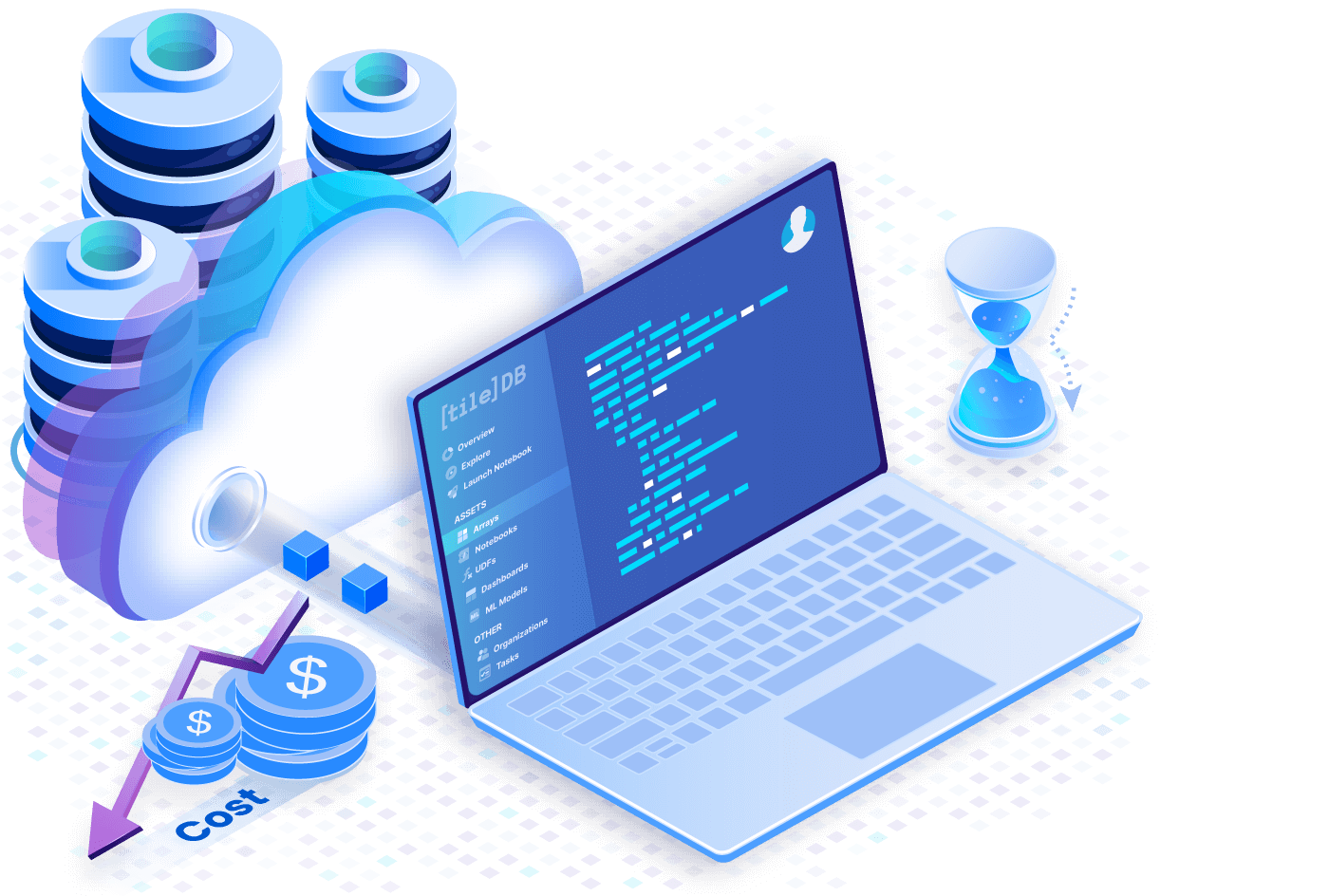
Harness the cloud
No more huge downloads and loading large datasets into memory. Cloud-native TileDB arrays allow you to slice straight from remote storage. Reduce costs and processing time by utilizing cost-efficient object storage services like S3.
Revolutionizing single-cell data analysis with TileDB Cloud
Model as analysis-ready arrays
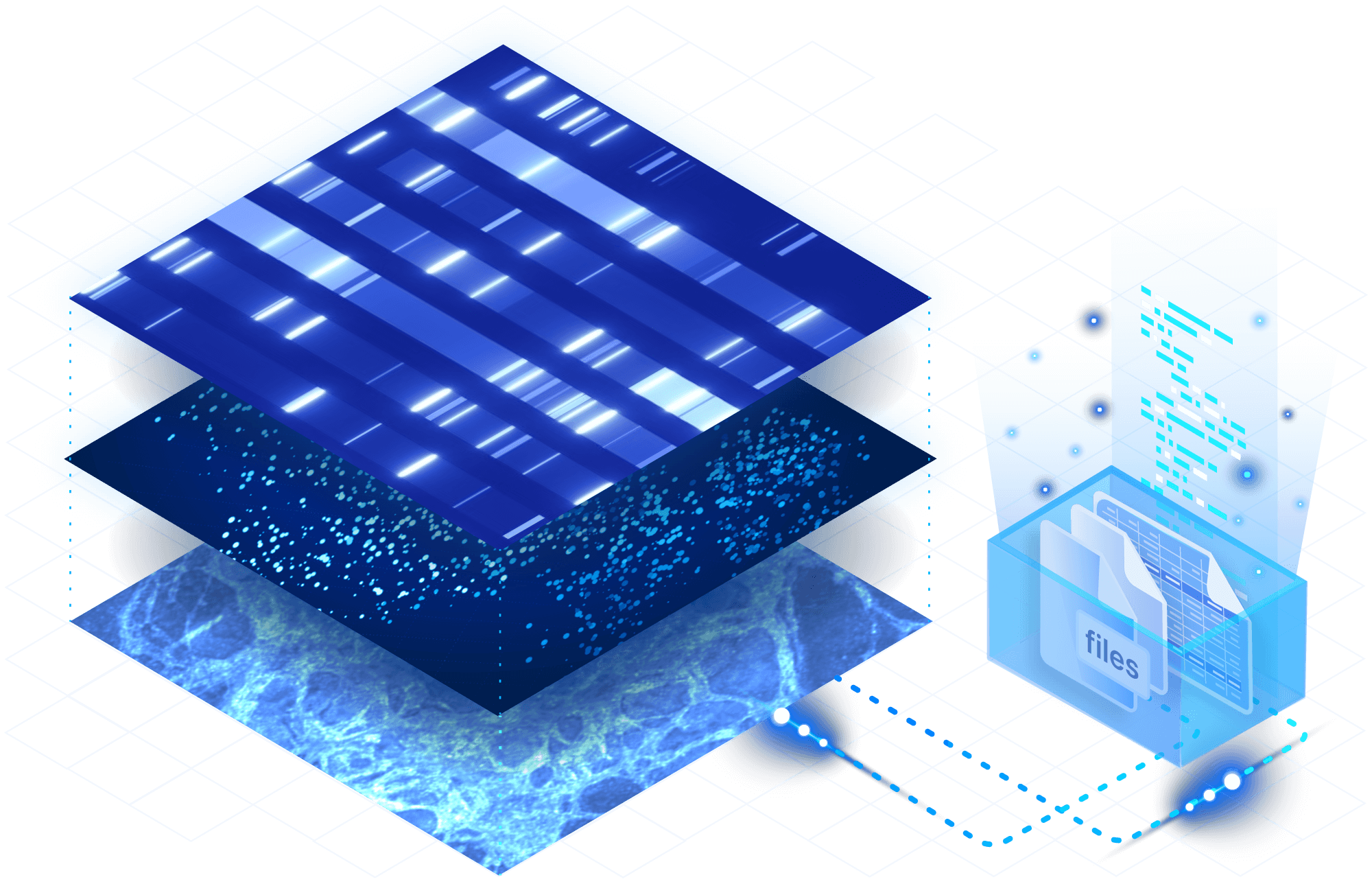
TileDB arrays are a natural fit for the highly sparse data generated by single-cell platforms. Manage RNA-seq data and count matrices in a cloud-native format, and store any number of samples in a compressed and lossless manner for tremendous storage savings.
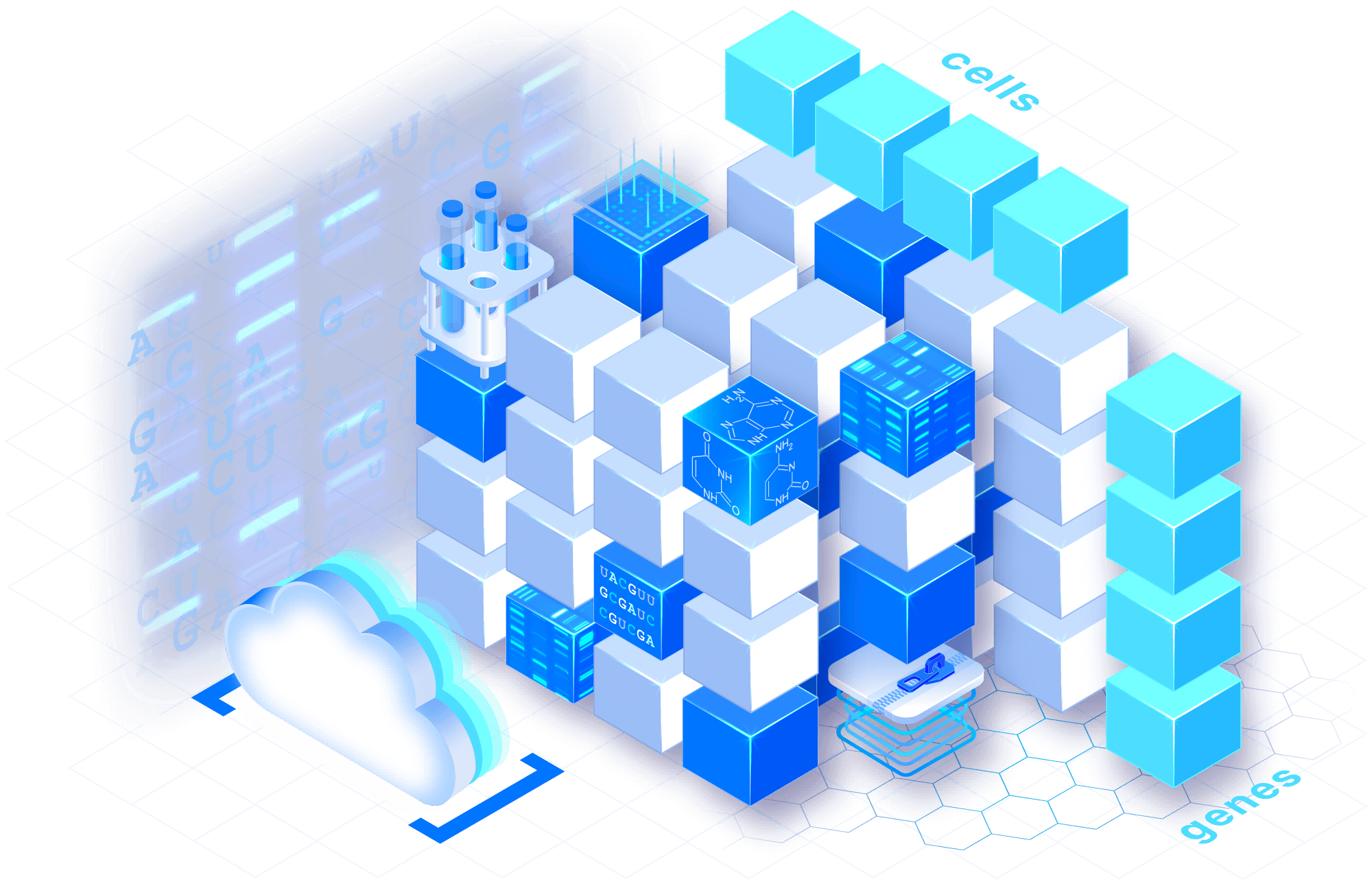
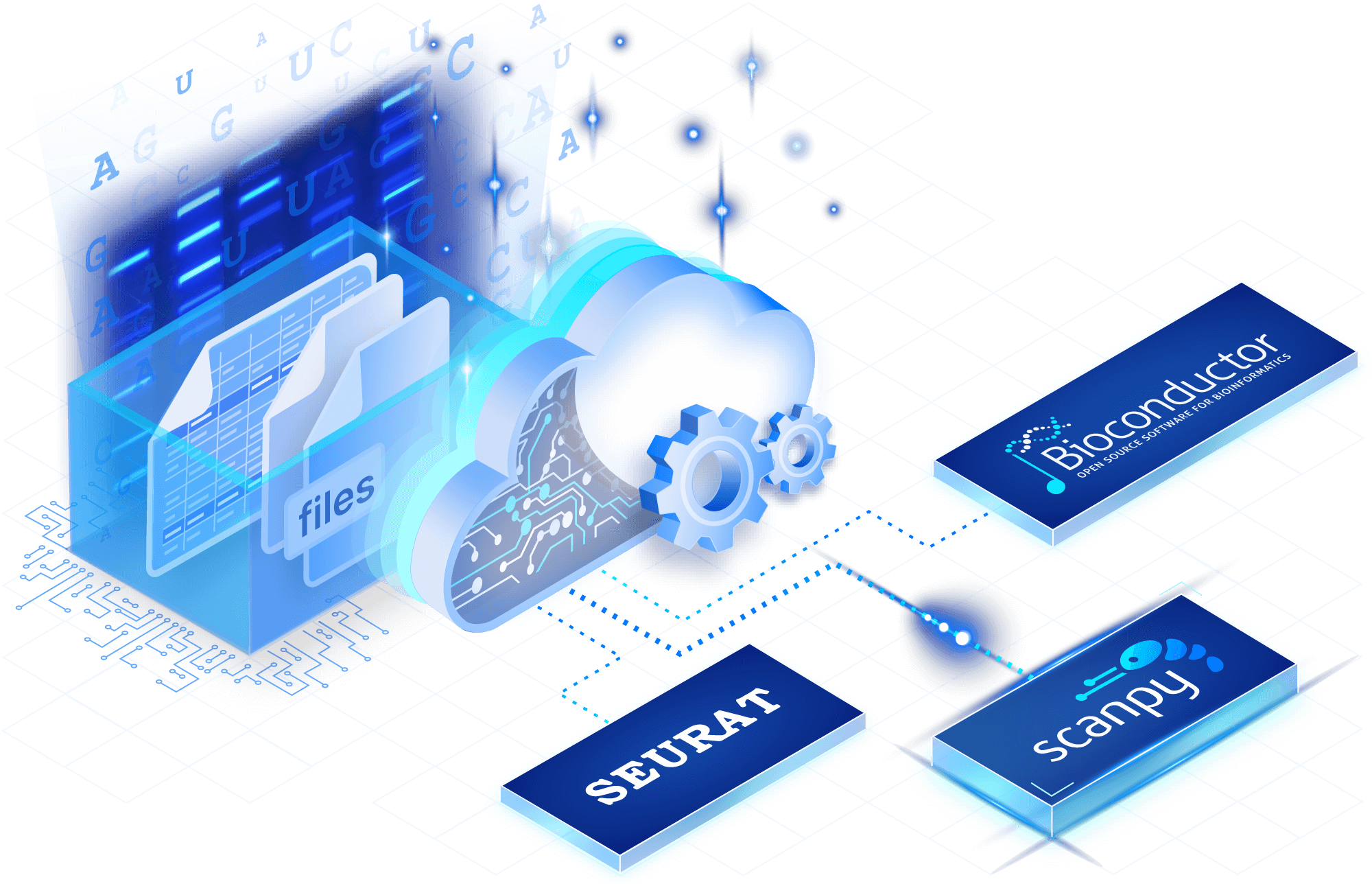
Use established tooling
Through our partnership with the Chan Zuckerberg Initiative on the Unified Single-Cell Data Model and API, TileDB implements a common, open data model. Use Seurat, Bioconductor and Scanpy, integrated directly with TileDB data.
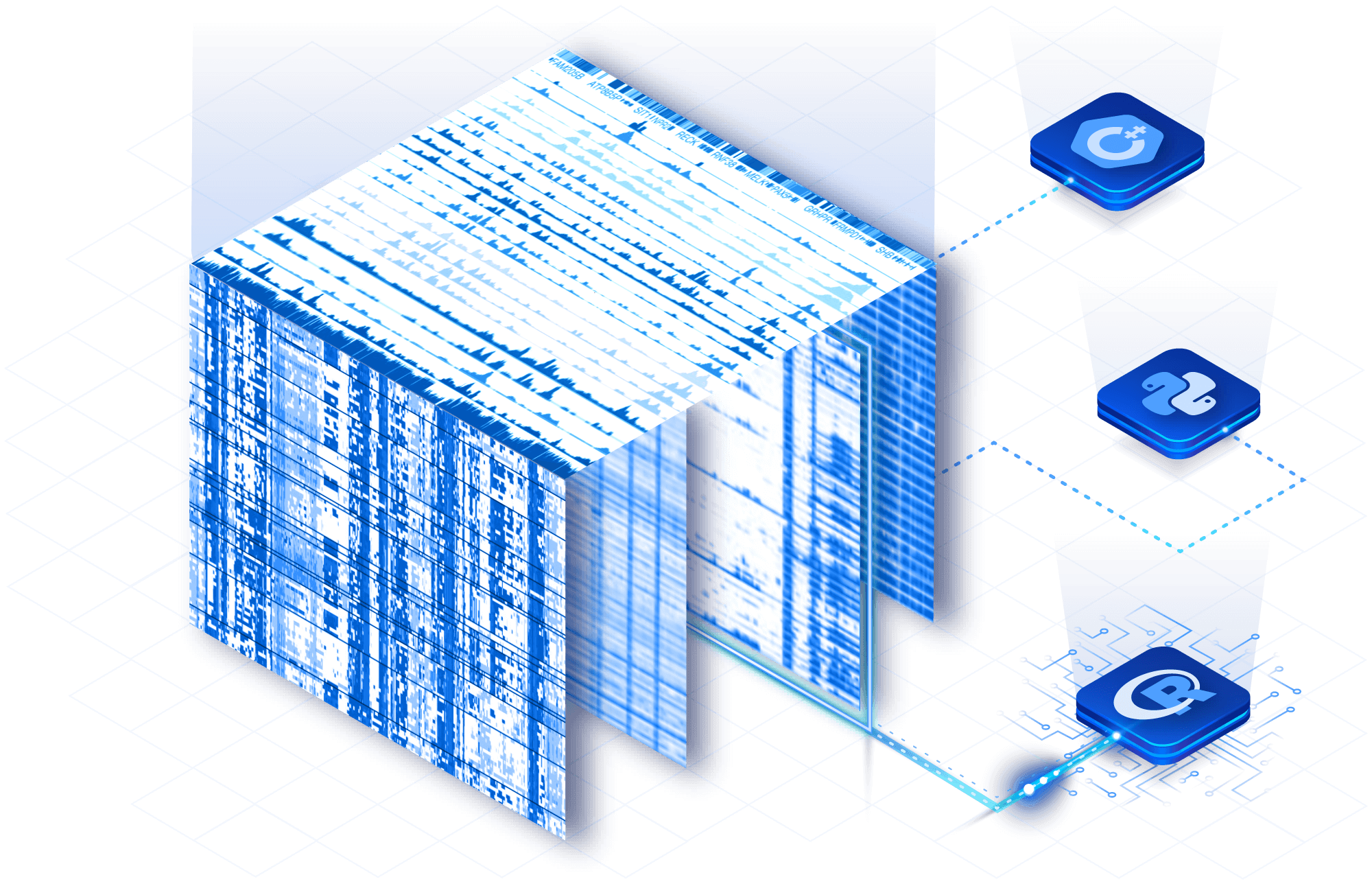
Slice directly by gene or cell
Eliminate data wrangling and large downloads. Slice TileDB arrays directly using native query conditions on genes and cells in any of TileDB’s language APIs, including R, Python and C++.
Combine modalities
Preserve spatial context. In addition to sparse data, the TileDB array format excels at representing dense data types like biomedical imaging. As your research grows, combine datasets, code, ML models, files and more into TileDB Cloud groups to manage large projects.
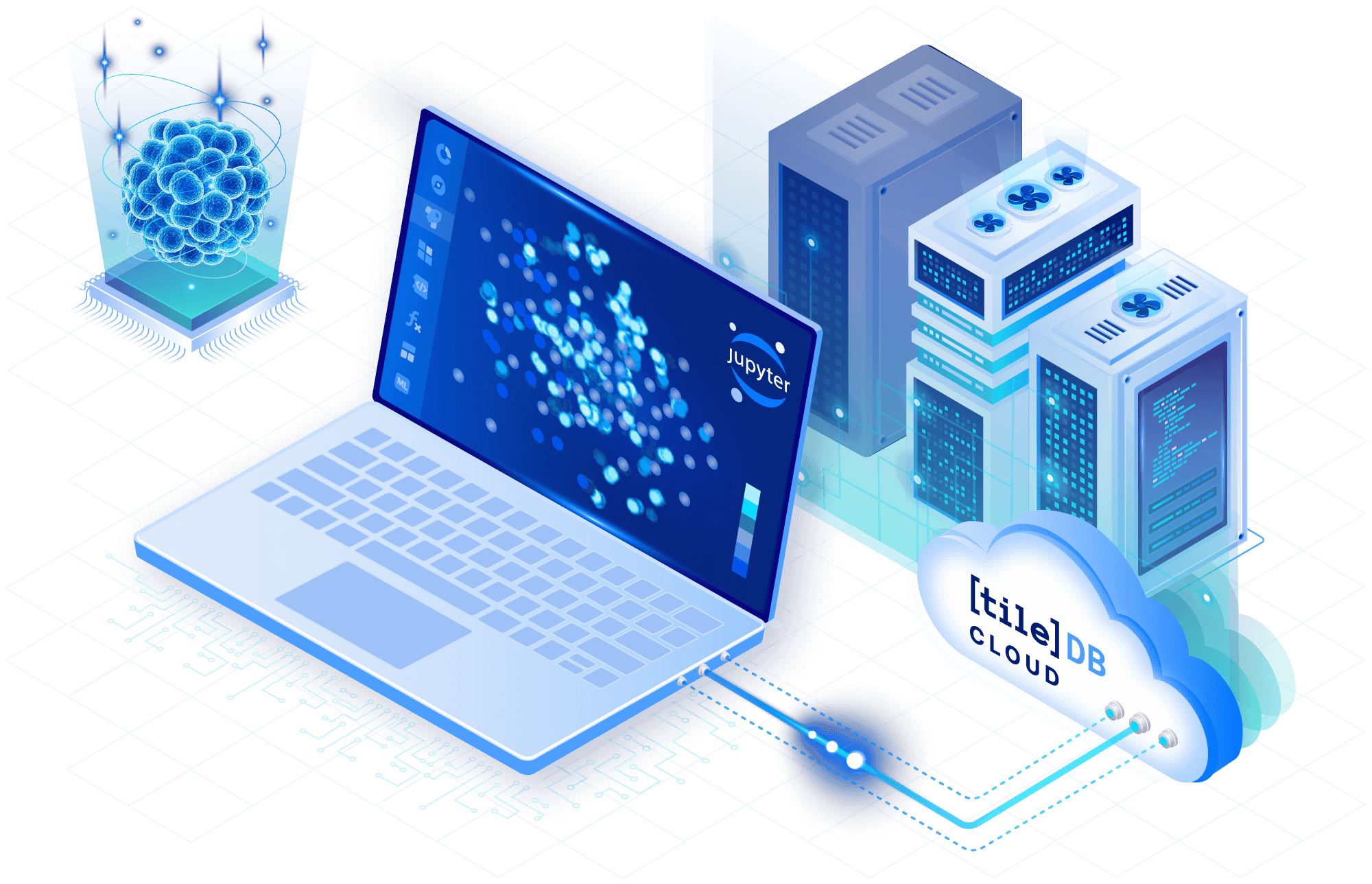
Run distributed analyses
Work across toolchains and languages to rapidly iterate using TileDB Cloud’s hosted Jupyter environment. For large computations, serverless user-defined functions and task graphs in TileDB Cloud let you parallelize custom algorithms efficiently and cost-effectively.
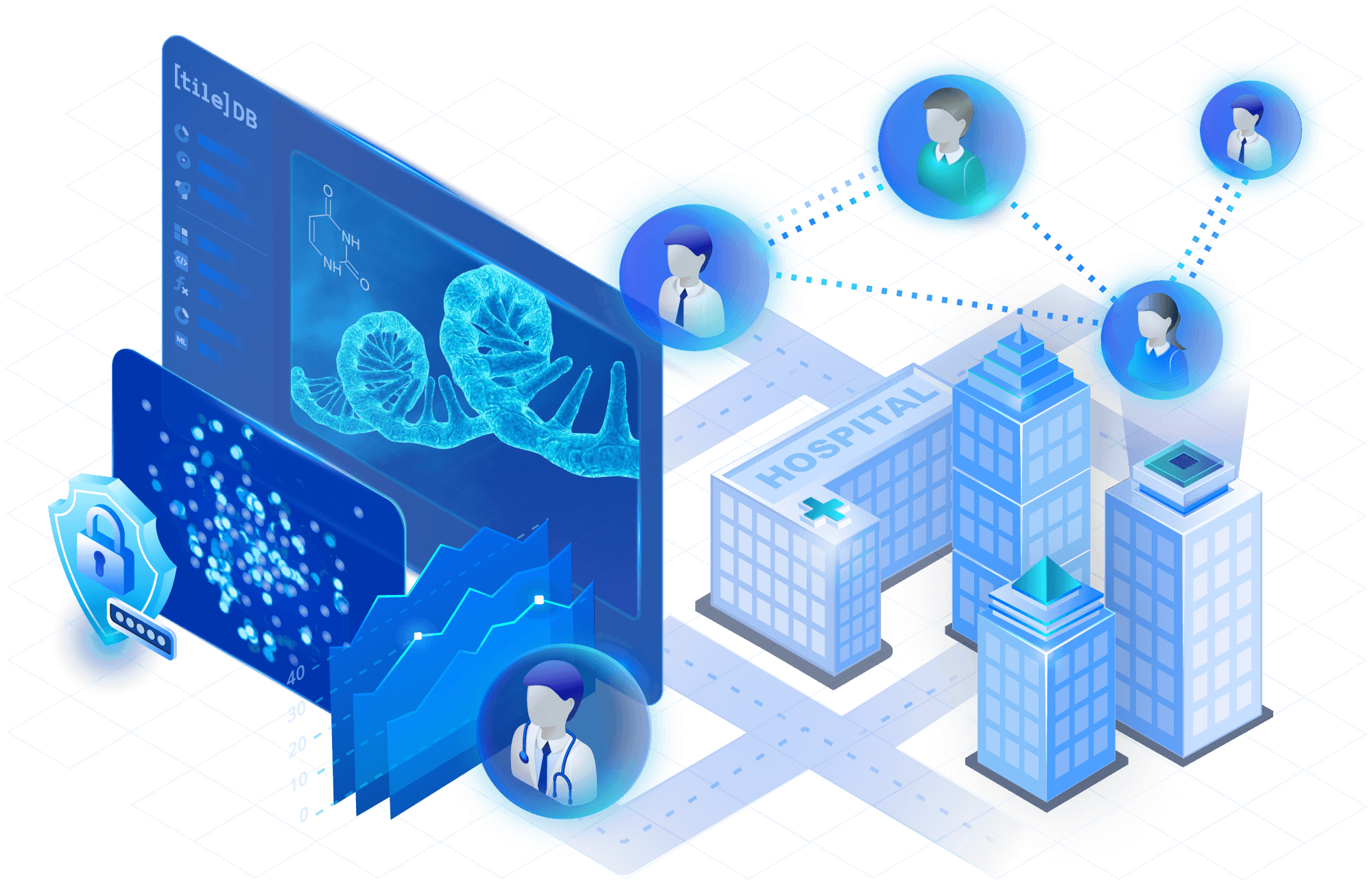
Securely distribute your repositories
Create your own repository of single-cell datasets using TileDB Cloud. Make cloud datasets discoverable with cataloging features that help attract new collaborators. Securely share data in private, or make it public in a predictable model that eliminates surprise infrastructure bills.
Open source code
We partnered with CZI to deliver the spec of an open source data model and corresponding implementations in R and Python for single-cell biology.
SOMA SPEC
SOMA is a flexible, extensible, and open-source data model for representing annotated matrices, commonly used in single cell biology.
Spec
TILEDB SOMA
TileDB-based implementations of the SOMA data model that allows you to slice data directly from the cloud into Seurat or Bioconductor in R, and AnnData in Python.
TileDB SOMA
Secret Sauce
Arrays
EMBEDDED
The open-source library that powers the open-source TileDB-SingleCell repository.
Learn moreDatabase
CLOUD
An advanced governance and computational platform for scaling your single-cell data and workloads.
Learn moreStay connected
Get product and feature updates.
Loading form...
Your personal data will be processed in accordance with TileDB's Privacy Policy.By subscribing you agree with TileDB, Inc. Terms of use. Your personal data will be processed in accordance with TileDB's Privacy Policy.