Population genomics data management made fast, cost-efficient and practical
Analyze and share enormous variant datasets with TileDB Cloud.
Variant data management made simple
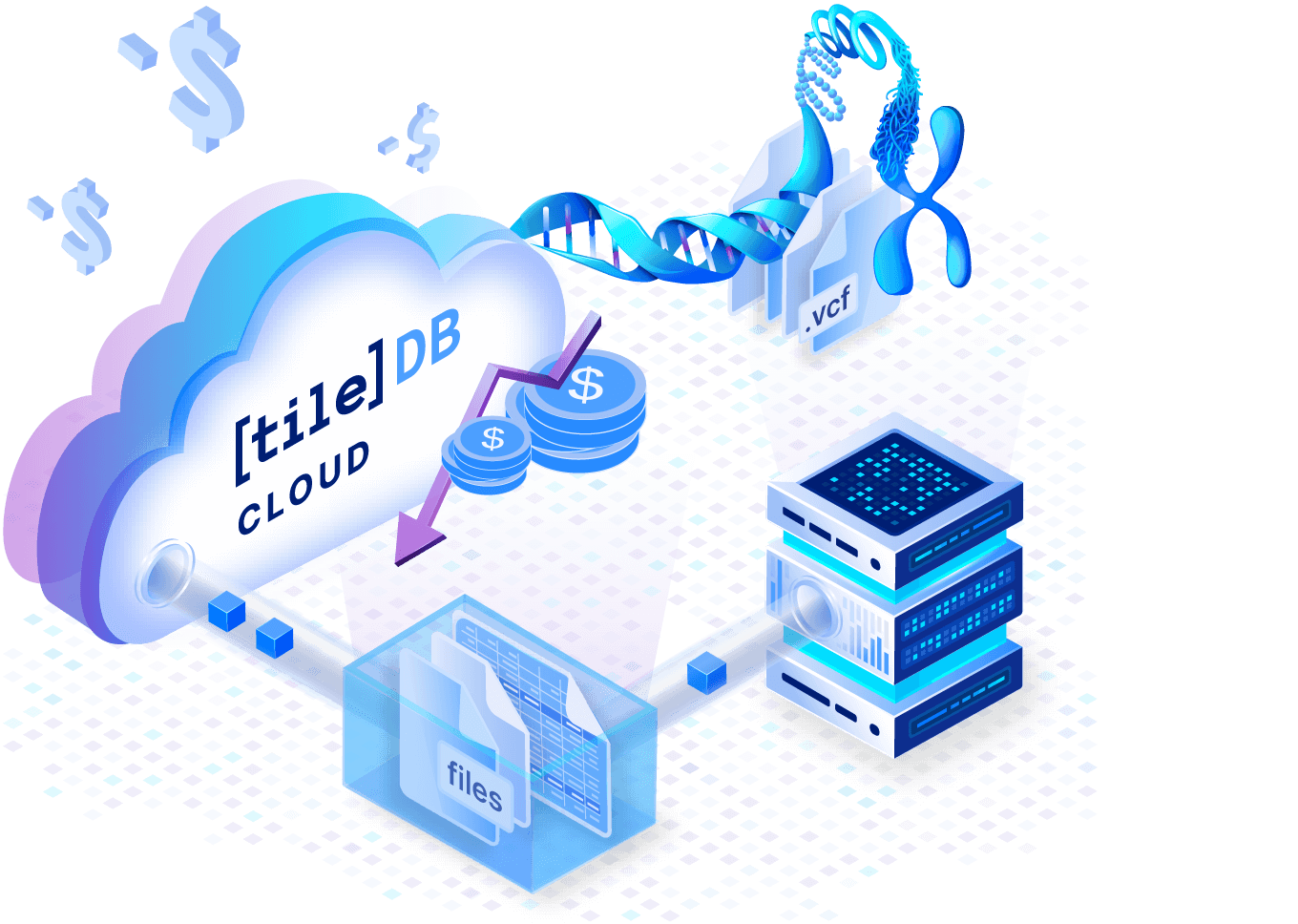
Performant and cost-effective ingest
Ingest data in parallel for as low as $0.01/per sample, for millions of samples. Rapidly add samples and refresh cohort summary statistics for billions of variants.
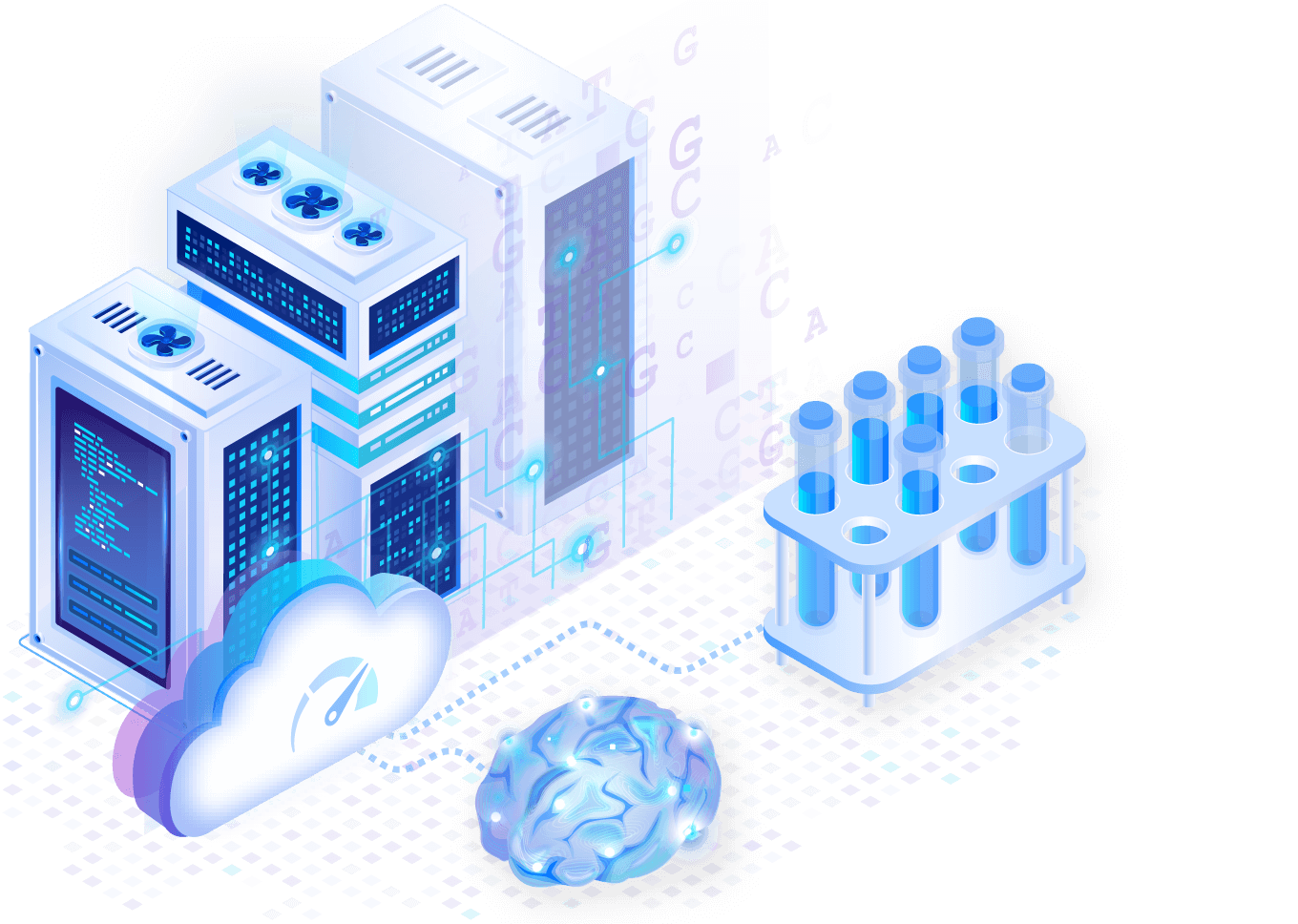
Genomic analysis at extreme scale
Run scalable pipelines without complex infrastructure hassles. Perform dynamic queries that integrate genome, phenome and clinical EHR data — all on a single platform.
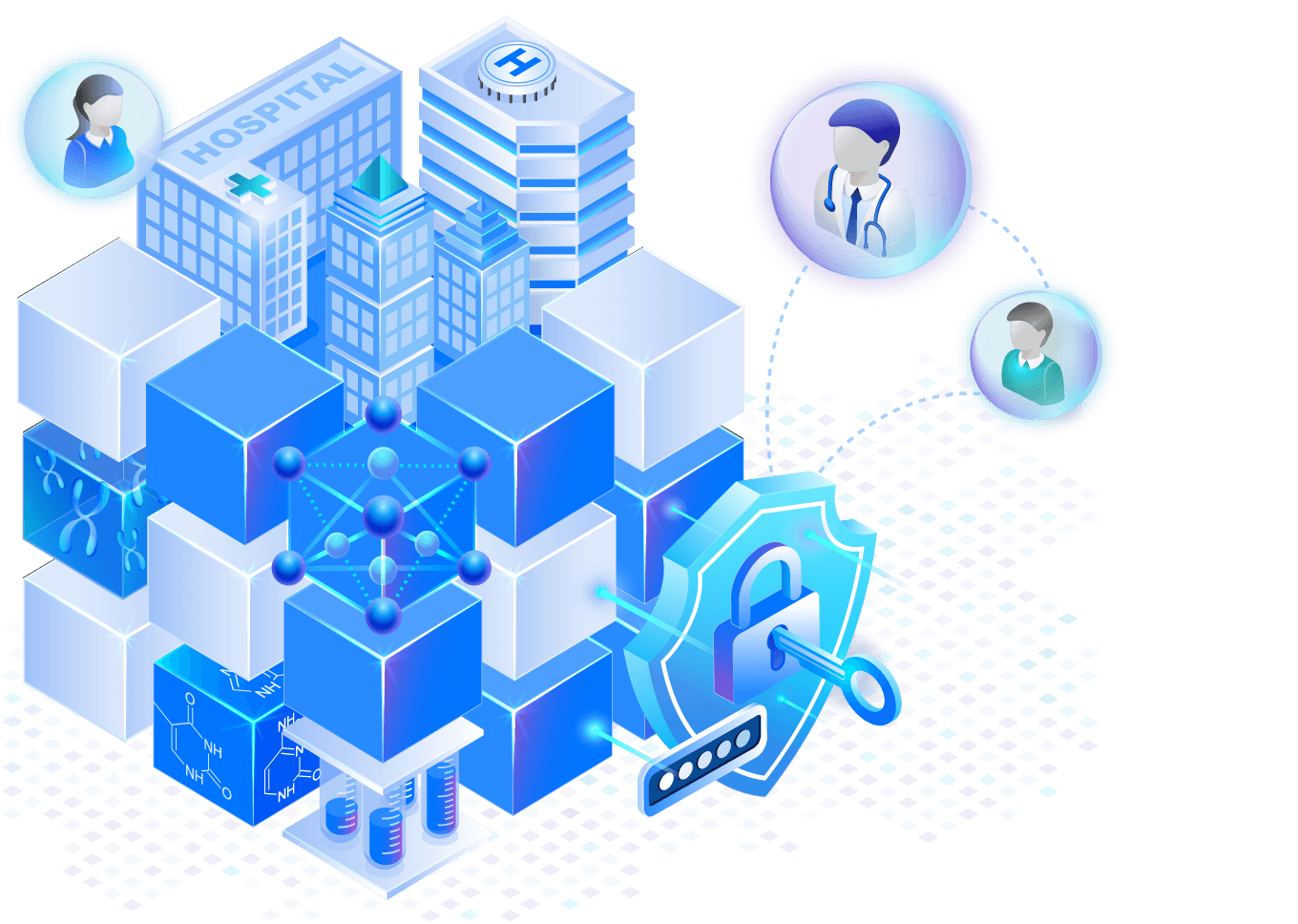
Governed, reproducible science
Deliver auditable and governed access to genomic datasets and code, enabling efficient collaboration with researchers and downstream partner organizations.
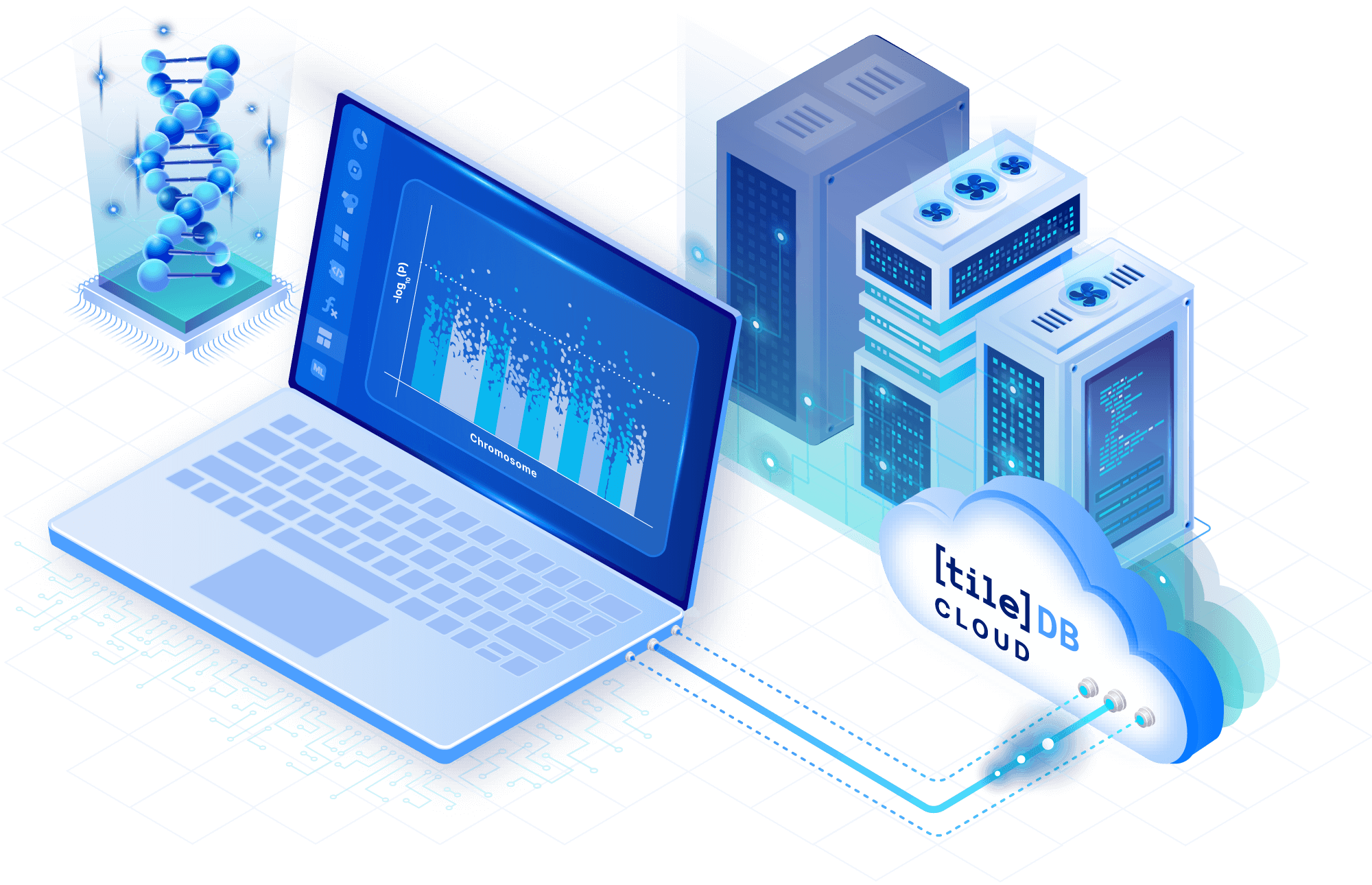
We reimagined genomics data management with TileDB Cloud
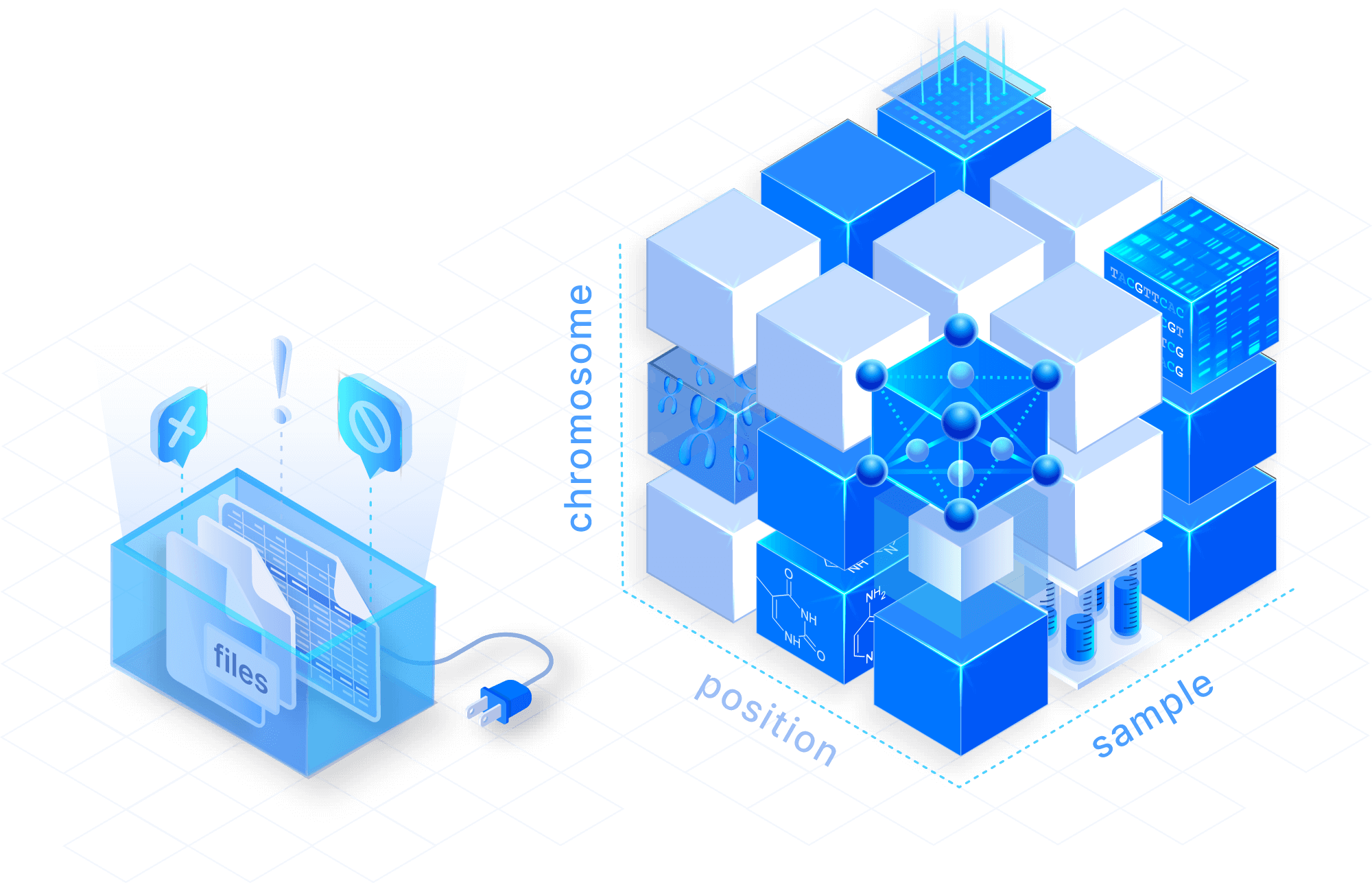
Model VCF data as analysis-ready arrays
Replace collections of VCF files with sparse multi-dimensional arrays. Manage variant data in a cloud-native format, and store any number of samples in a compressed and lossless manner for tremendous storage savings.
Ingest massive datasets in parallel
TileDB supports parallel writes enabling ingestion to be scaled out across multiple nodes with zero inter-process communication. VCF parsing is split across multiple workers and remote files are downloaded asynchronously.
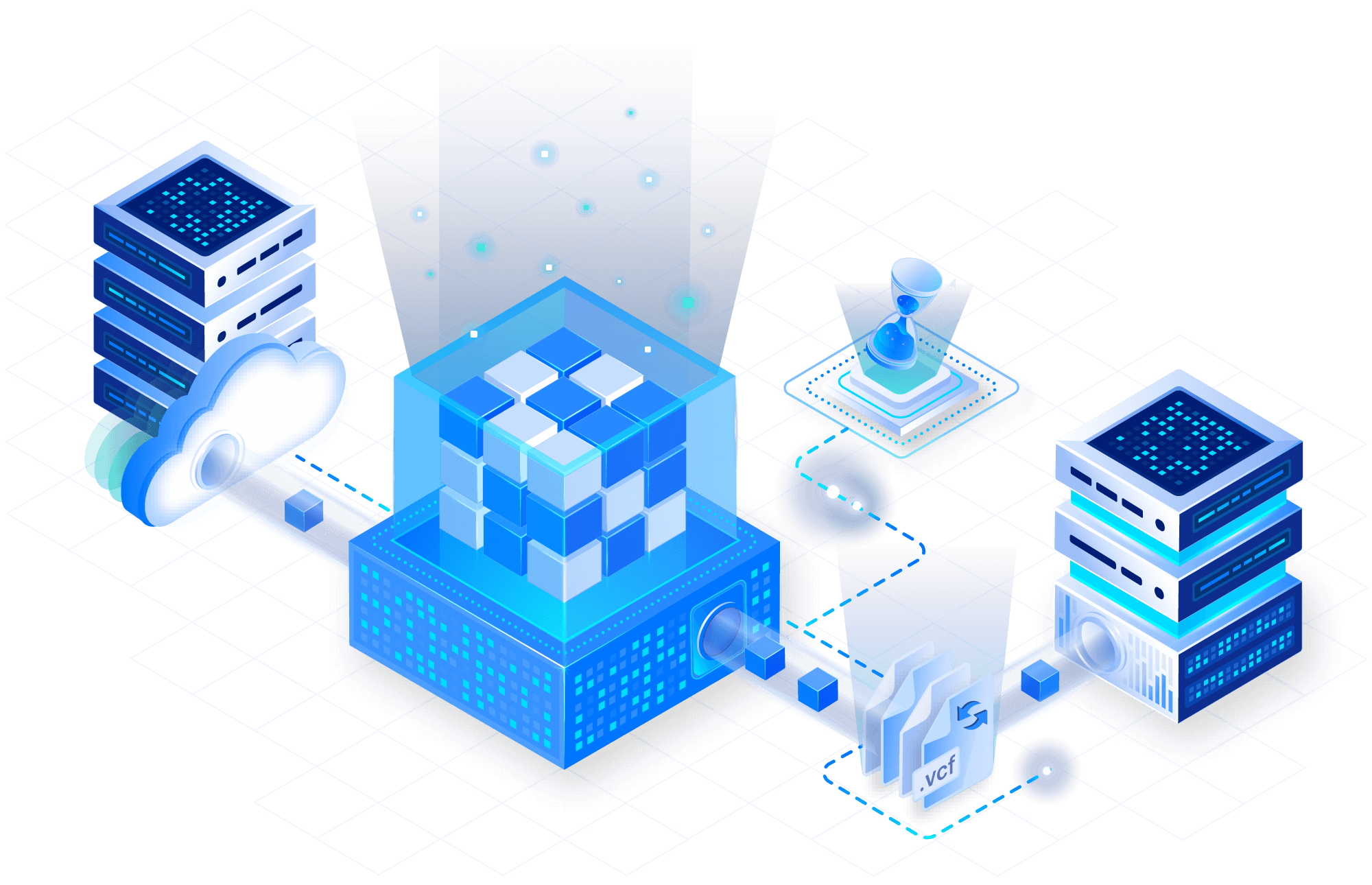
Solve the N+1 problem
TileDB arrays are updatable with arbitrary metadata and versioning built-in. Rapidly add new samples, scaling storage linearly. Avoid unnecessary storage inflation by maintaining the natively sparse format of the variant data.
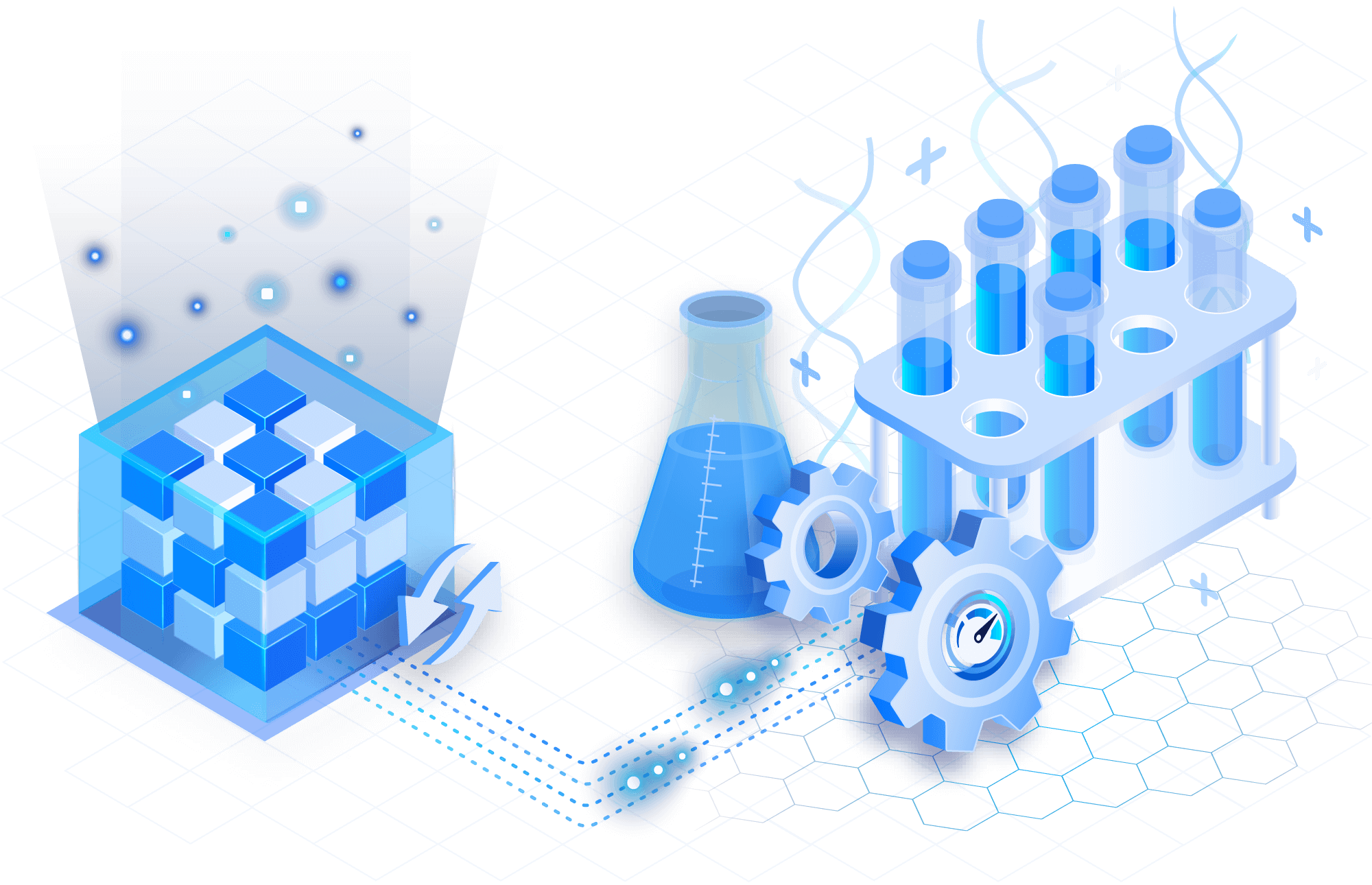
Slice and filter in real time
Efficiently explore any VCF field such as chromosome, position or sample. Leverage a dedicated C++ library that is optimized for querying variant records by genomic regions across arbitrary numbers of samples.
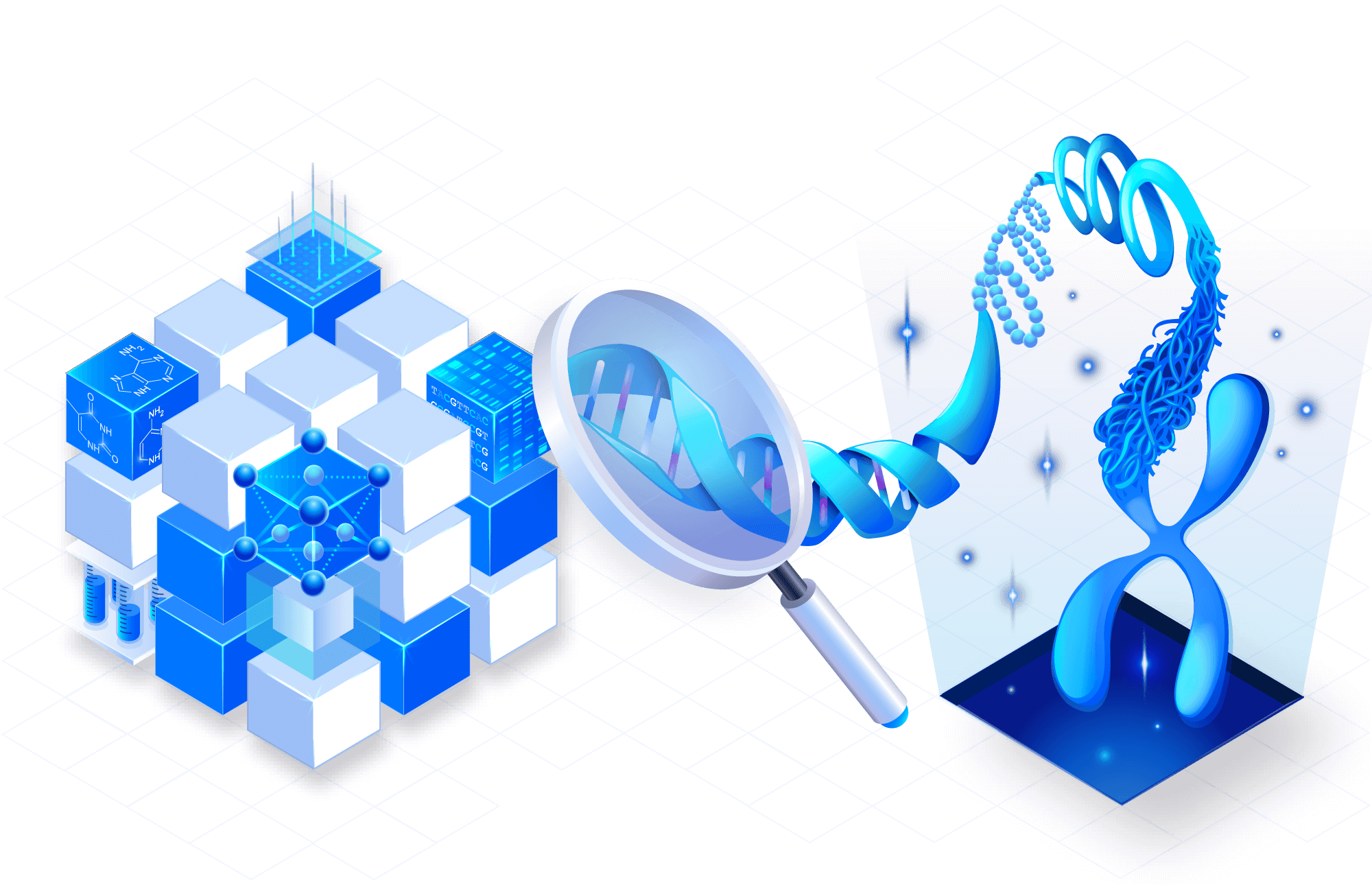
Securely govern your data
TileDB Cloud is designed from the ground-up with strict governance and auditing guarantees. Access control on each dataset enables easy, auditable sharing for individual users or entire organizations. No more expensive data rehosting costs and cumbersome downloads.
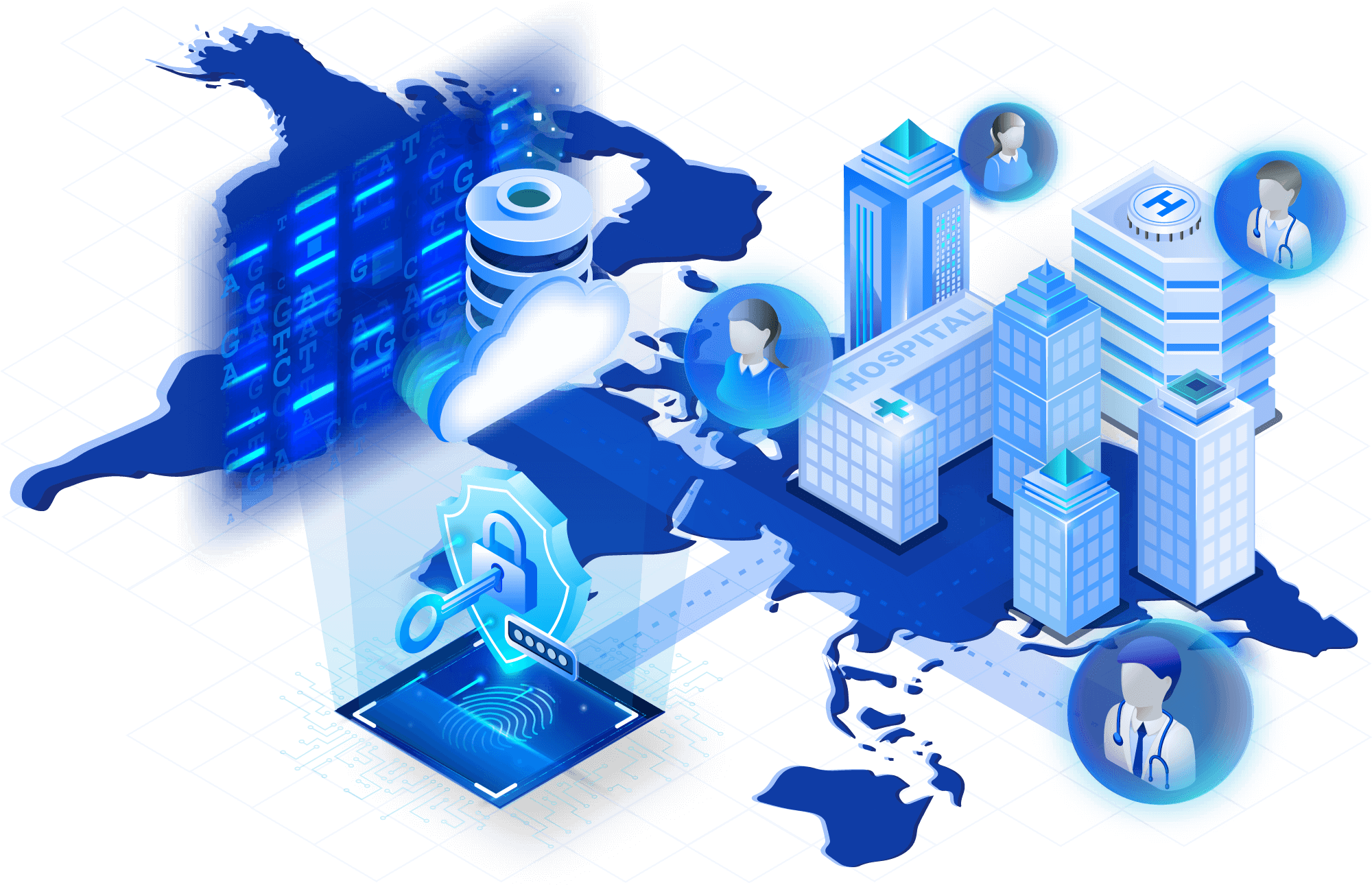
Eliminate cluster hassles
TileDB's serverless design allows you to scale workloads without manually spinning up clusters. Run pipelines, perform genome-wide analyses and calculate sample- or variant-level metrics on TileDB Cloud, right from your browser.
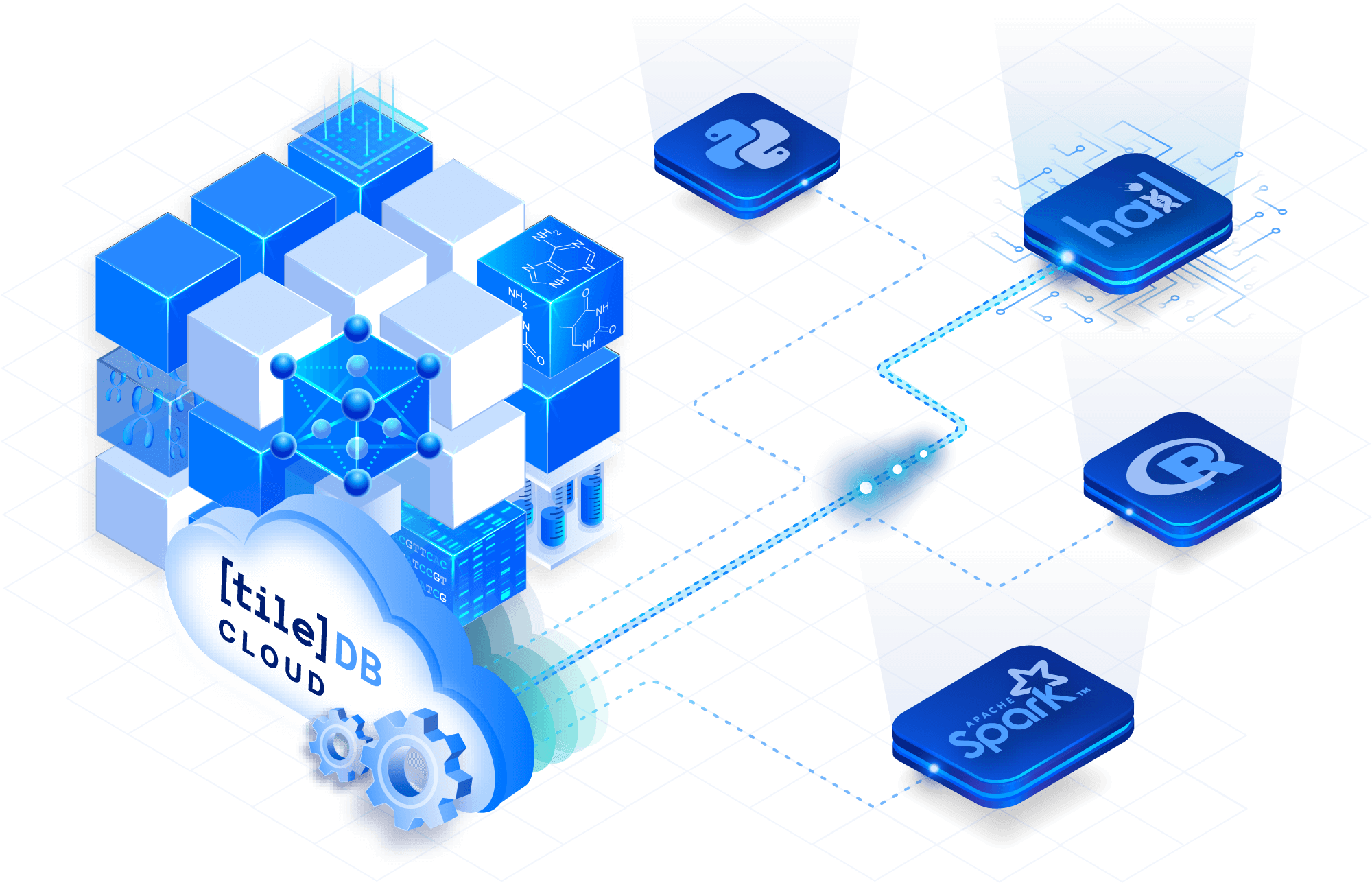
Interoperate with popular tools
Access variant data with a variety of APIs and integrations, including Python, R, Java, Spark and Hail. You can also losslessly export to the original VCF files, as well as to combined VCF files, providing compatibility with popular tools for genomic analysis.
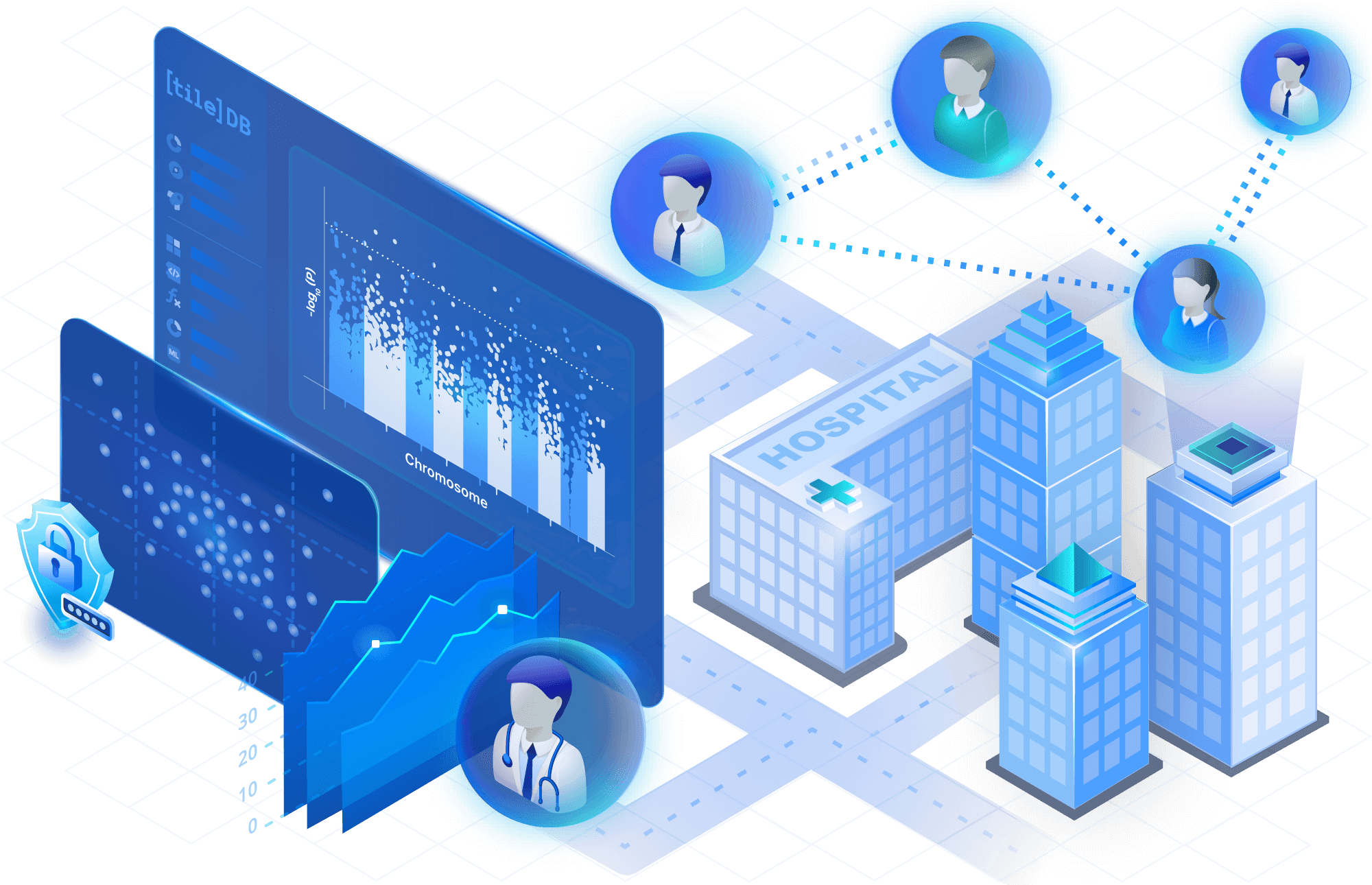
Collaborate easily
Share large datasets, notebooks, code, even dashboards, within and outside your organization. Eliminate downloads and any kind of data movement. Log and audit every action on your data and code assets.
Secret Sauce
Resources
Documentation
Have a technical question? Check out our comprehensive documentation, learn about our genomics integrations and dig into API reference materials.
Read the docsWebinar
Watch the replay! Aaron Wolen, Senior Software Engineer, presented code examples, from data modeling to sharing access to multi-TB variant datasets.
Watch the videoTutorial
Ready to get started? Jump right in with the TileDB VCF Quickstart tutorial on TileDB Cloud using Jupyter notebooks directly through its console.
Go to tutorialStay connected
Get product and feature updates.
Loading form...
Your personal data will be processed in accordance with TileDB's Privacy Policy.By subscribing you agree with TileDB, Inc. Terms of use. Your personal data will be processed in accordance with TileDB's Privacy Policy.